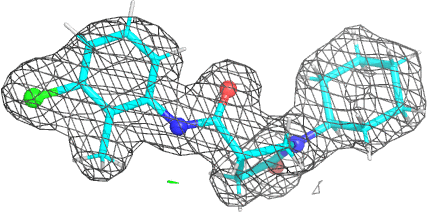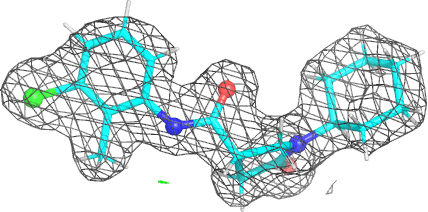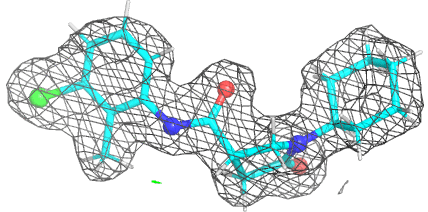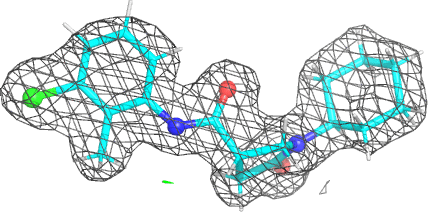
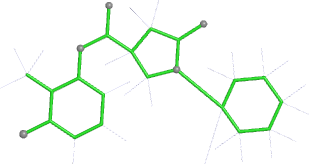
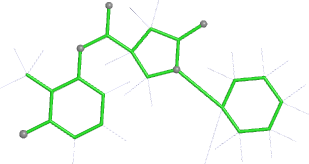
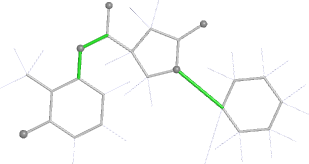
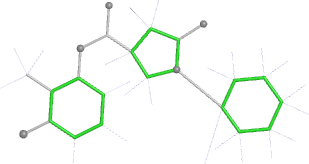
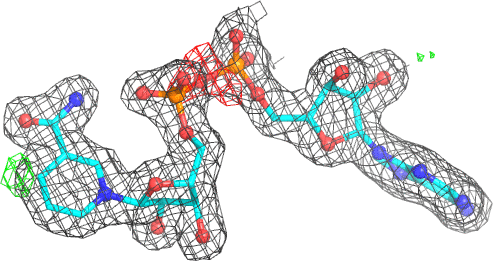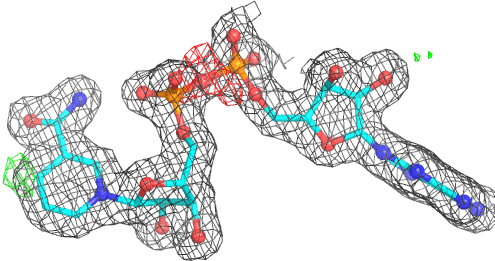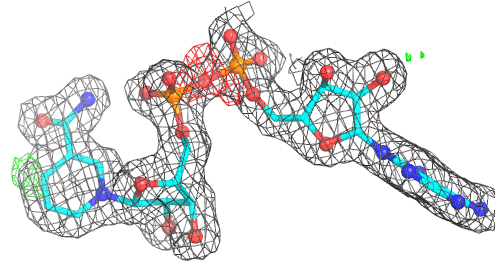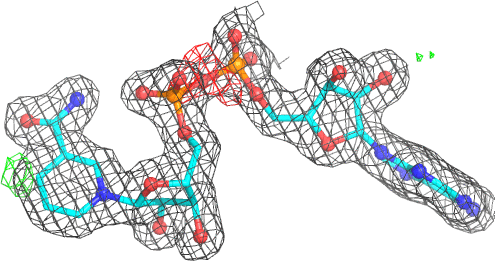
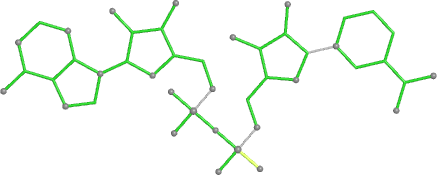
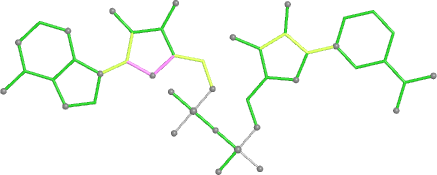
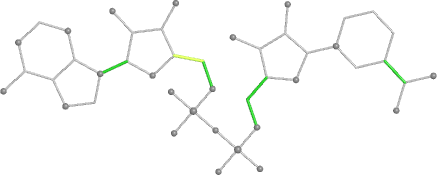
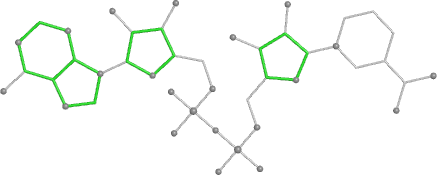
A 501
Electron density around 468 A 501 :
2mFo-DFc (at 0.7 rmsd) in grey.
mFo-DFc (at 3 rmsd) in red (negative) and green (positive),
(picture produced with PyMOL)
| Database ID | 468 (PDB) |
| 3-letter code | 468 |
| CC(2mFo-DFc) | 0.964 |
| min(B-factor)‡ | 26.3 |
| avg(B-factor)‡ | 32.7 |
| max(B-factor)‡ | 54.8 |
| min(occupancy)‡ | 1.00 |
| max(occupancy)‡ | 1.00 |
| ‡ hydrogen atoms excluded | |
| Mogul Analysis: | |
|---|---|
| 'bad' bonds | 0/25 |
| 'bad' bond angles | 0/31 |
| 'unusual' dihedrals | 0/3 |
| 'bad' rings | 0/3 |
| bonds rms Z | 0.3 |
| angles rms Z | 0.5 |
Restraints used
Detailed report
help?
Mogul bonds | Mogul angles |
Mogul dihedrals |
Mogul rings |
A 500
Electron density around NAD A 500 :
2mFo-DFc (at 0.7 rmsd) in grey.
mFo-DFc (at 3 rmsd) in red (negative) and green (positive),
(picture produced with PyMOL)
| Database ID | NAD (PDB) |
| 3-letter code | NAD |
| CC(2mFo-DFc) | 0.981 |
| min(B-factor)‡ | 17.7 |
| avg(B-factor)‡ | 22.0 |
| max(B-factor)‡ | 25.5 |
| min(occupancy)‡ | 1.00 |
| max(occupancy)‡ | 1.00 |
| ‡ hydrogen atoms excluded | |
| Mogul Analysis: | |
|---|---|
| 'bad' bonds | 0/45 |
| 'bad' bond angles | 0/60 |
| 'unusual' dihedrals | 0/6 |
| 'bad' rings | 0/4 |
| bonds rms Z | 0.6 |
| angles rms Z | 0.9 |
Restraints used
Detailed report
help?
Mogul bonds | Mogul angles |
Mogul dihedrals |
Mogul rings |