Electron density around 468 A 501 :
2mFo-DFc (at 0.7 rmsd) in grey.
mFo-DFc (at 3 rmsd) in red (negative) and green (positive),
(picture produced with PyMOL)
Electron density around 468 A 501 : |
| Database ID | 468 (PDB) |
| 3-letter code | 468 |
| CC(2mFo-DFc) | 0.9643 |
| min(B-factor)‡ | 26.3 |
| avg(B-factor)‡ | 32.7 |
| max(B-factor)‡ | 54.8 |
| min(occupancy)‡ | 1.00 |
| max(occupancy)‡ | 1.00 |
| ‡ hydrogen atoms excluded | |
| Mogul Analysis: | |
|---|---|
| 'bad' bonds | 0/25 |
| 'bad' bond angles | 0/31 |
| 'unusual' dihedrals | 0/3 |
| 'bad' rings | 0/3 |
| bonds rms Z | 0.282 |
| angles rms Z | 0.486 |
Mogul bonds schematic | ||||||
| atoms | actual in Å | Mogul mean in Å | difference in Å | Mogul σ in Å | Mogul # samples | Zscore |
|---|---|---|---|---|---|---|
| C20-C15 | 1.400 | 1.392 | 0.008 | 0.012 | 3256 | 0.73 |
| C3-N11 | 1.477 | 1.470 | 0.007 | 0.013 | 21 | 0.51 |
| C6-C5 | 1.525 | 1.516 | 0.009 | 0.018 | 1743 | 0.51 |
| C10-C9 | 1.539 | 1.530 | 0.009 | 0.019 | 36 | 0.45 |
| C15-C16 | 1.405 | 1.399 | 0.006 | 0.013 | 49 | 0.44 |
| C2-C3 | 1.527 | 1.522 | 0.006 | 0.014 | 1191 | 0.39 |
| C18-C17 | 1.379 | 1.384 | -0.005 | 0.013 | 2673 | 0.34 |
| C6-C1 | 1.521 | 1.516 | 0.005 | 0.018 | 1743 | 0.30 |
| C17-C16 | 1.393 | 1.391 | 0.002 | 0.008 | 28 | 0.26 |
| C8-C9 | 1.527 | 1.531 | -0.004 | 0.021 | 57 | 0.20 |
| C19-C18 | 1.383 | 1.384 | -0.002 | 0.010 | 2659 | 0.16 |
| C17-CL1 | 1.732 | 1.734 | -0.002 | 0.012 | 3693 | 0.15 |
| C5-C4 | 1.526 | 1.525 | 0.001 | 0.013 | 1827 | 0.08 |
| C9-C12 | 1.511 | 1.509 | 0.002 | 0.019 | 46 | 0.08 |
| C23-C16 | 1.508 | 1.509 | -0.001 | 0.011 | 1974 | 0.07 |
| C1-C2 | 1.526 | 1.525 | 0.001 | 0.013 | 1827 | 0.06 |
| C12-N13 | 1.351 | 1.350 | 0.001 | 0.014 | 362 | 0.05 |
| O15-C7 | 1.220 | 1.220 | -0.000 | 0.014 | 2954 | 0.04 |
| C19-C20 | 1.384 | 1.384 | -0.000 | 0.010 | 2659 | 0.04 |
| C7-N11 | 1.345 | 1.346 | -0.001 | 0.013 | 191 | 0.04 |
| C4-C3 | 1.522 | 1.522 | 0.000 | 0.014 | 1191 | 0.03 |
| O14-C12 | 1.228 | 1.228 | 0.000 | 0.012 | 3021 | 0.03 |
| C15-N13 | 1.414 | 1.415 | -0.000 | 0.014 | 1552 | 0.02 |
| C8-C7 | 1.505 | 1.505 | 0.000 | 0.010 | 228 | 0.01 |
| C10-N11 | 1.465 | 1.465 | 0.000 | 0.013 | 181 | 0.01 |
Mogul angles schematic | ||||||
| atoms | actual in ° | Mogul mean in ° | difference in ° | Mogul σ in ° | Mogul # samples | Zscore |
|---|---|---|---|---|---|---|
| C8-C9-C12 | 109.4 | 112.1 | -2.7 | 1.8 | 16 | 1.47 |
| C18-C17-CL1 | 117.0 | 118.4 | -1.4 | 1.3 | 2376 | 1.02 |
| C10-N11-C3 | 122.0 | 124.2 | -2.2 | 2.4 | 11 | 0.90 |
| C3-N11-C7 | 124.5 | 122.7 | 1.8 | 2.1 | 16 | 0.85 |
| C20-C15-N13 | 120.3 | 121.9 | -1.6 | 2.2 | 1192 | 0.76 |
| C16-C15-N13 | 118.4 | 119.3 | -0.9 | 1.6 | 18 | 0.58 |
| C4-C3-C2 | 110.1 | 111.0 | -0.9 | 1.7 | 435 | 0.51 |
| O14-C12-C9 | 121.6 | 122.1 | -0.5 | 1.0 | 39 | 0.49 |
| C1-C2-C3 | 110.2 | 110.9 | -0.7 | 1.4 | 901 | 0.46 |
| O14-C12-N13 | 123.7 | 124.2 | -0.5 | 1.2 | 305 | 0.44 |
| C19-C20-C15 | 119.6 | 119.0 | 0.6 | 1.7 | 2995 | 0.36 |
| C9-C12-N13 | 114.7 | 114.8 | -0.2 | 0.6 | 11 | 0.30 |
| C10-N11-C7 | 113.5 | 113.8 | -0.3 | 1.0 | 14 | 0.29 |
| C5-C4-C3 | 110.5 | 110.9 | -0.4 | 1.4 | 901 | 0.28 |
| C18-C19-C20 | 120.2 | 120.4 | -0.3 | 1.0 | 1853 | 0.27 |
| C4-C3-N11 | 111.5 | 111.7 | -0.2 | 0.9 | 18 | 0.22 |
| C23-C16-C17 | 122.4 | 122.2 | 0.2 | 0.9 | 26 | 0.22 |
| C5-C6-C1 | 111.1 | 110.9 | 0.2 | 1.0 | 1269 | 0.20 |
| C18-C17-C16 | 122.9 | 123.0 | -0.1 | 0.5 | 22 | 0.20 |
| C6-C1-C2 | 111.4 | 111.3 | 0.2 | 1.0 | 1421 | 0.19 |
| C19-C18-C17 | 119.3 | 119.4 | -0.1 | 0.7 | 1865 | 0.18 |
| O15-C7-C8 | 127.0 | 127.3 | -0.2 | 1.1 | 174 | 0.18 |
| C2-C3-N11 | 111.8 | 111.7 | 0.1 | 0.9 | 18 | 0.15 |
| O15-C7-N11 | 124.7 | 124.8 | -0.1 | 1.1 | 162 | 0.10 |
| C6-C5-C4 | 111.2 | 111.3 | -0.1 | 1.0 | 1421 | 0.07 |
| C15-N13-C12 | 126.3 | 126.5 | -0.1 | 2.4 | 92 | 0.06 |
| C9-C10-N11 | 102.0 | 102.1 | -0.1 | 1.8 | 10 | 0.05 |
| C20-C15-C16 | 121.3 | 121.3 | 0.0 | 0.8 | 35 | 0.05 |
| C23-C16-C15 | 121.2 | 121.2 | 0.0 | 1.0 | 42 | 0.04 |
| C8-C7-N11 | 108.2 | 108.2 | 0.0 | 1.0 | 22 | 0.02 |
| C16-C17-CL1 | 119.5 | 119.6 | -0.0 | 0.6 | 26 | 0.01 |
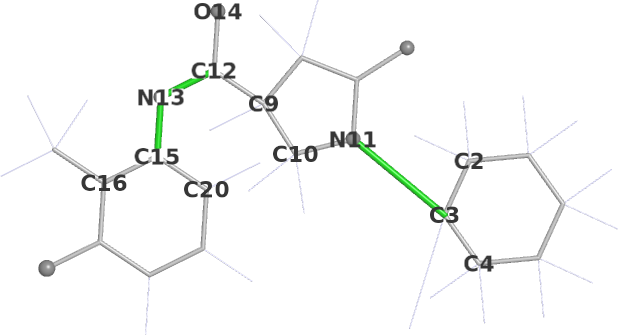 Mogul dihedrals schematic | ||||
| atoms | actual torsion angle in ° | Mogul histogram | Mogul # samples | classification or % Mogul population within ± 10° |
|---|---|---|---|---|
| …-C12-N13-… | common | |||
| O14-C12-N13-C15 | 3.7 |  | 340 | 99% |
| C9-C12-N13-C15 | -175.2 |  | 87 | 100% |
| …-C15-N13-… | common | |||
| C16-C15-N13-C12 | 146.5 | 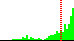 | 354 | 27% |
| C20-C15-N13-C12 | -32.1 | 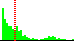 | 934 | 30% |
| …-C3-N11-… | common | |||
| C2-C3-N11-C10 | 36.5 | 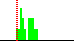 | 16 | 31% |
| C4-C3-N11-C10 | -87.3 | 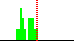 | 16 | 25% |
Mogul rings schematic | ||
| atoms | Mogul # samples | Ring strangeness score‡ in ° |
|---|---|---|
| C7-C8-C9-C10-N11 | 10 | 1.1 |
| C15-C20-C19-C18-C17-C16 | 167 | 1.1 |
| C1-C2-C3-C4-C5-C6 | 67 | 0.5 |
‡ 'ring strangeness score' is the RMS difference in torsion angles between the instance of the ring in the ligand in the model, and the nearest instance that mogul finds in the CSD.