Electron density around NAD A 500 :
2mFo-DFc (at 0.7 rmsd) in grey.
mFo-DFc (at 3 rmsd) in red (negative) and green (positive),
(picture produced with PyMOL)
Electron density around NAD A 500 : |
| Database ID | NAD (PDB) |
| 3-letter code | NAD |
| CC(2mFo-DFc) | 0.9812 |
| min(B-factor)‡ | 17.7 |
| avg(B-factor)‡ | 22.0 |
| max(B-factor)‡ | 25.5 |
| min(occupancy)‡ | 1.00 |
| max(occupancy)‡ | 1.00 |
| ‡ hydrogen atoms excluded | |
| Mogul Analysis: | |
|---|---|
| 'bad' bonds | 0/45 |
| 'bad' bond angles | 0/60 |
| 'unusual' dihedrals | 0/6 |
| 'bad' rings | 0/4 |
| bonds rms Z | 0.553 |
| angles rms Z | 0.907 |
Mogul bonds schematic | ||||||
| atoms | actual in Å | Mogul mean in Å | difference in Å | Mogul σ in Å | Mogul # samples | Zscore |
|---|---|---|---|---|---|---|
| PN-O2N | 1.481 | 1.527 | -0.046 | 0.030 | 30 | 1.54 |
| C3B-C4B | 1.543 | 1.526 | 0.018 | 0.014 | 577 | 1.28 |
| PA-O2A | 1.489 | 1.527 | -0.038 | 0.030 | 30 | 1.28 |
| O4D-C4D | 1.455 | 1.444 | 0.011 | 0.011 | 1633 | 1.01 |
| C2B-C1B | 1.518 | 1.530 | -0.013 | 0.014 | 341 | 0.90 |
| C4N-C3N | 1.401 | 1.391 | 0.011 | 0.013 | 4266 | 0.84 |
| O3D-C3D | 1.434 | 1.423 | 0.010 | 0.013 | 3264 | 0.79 |
| C7N-N7N | 1.344 | 1.327 | 0.016 | 0.021 | 568 | 0.79 |
| C6N-N1N | 1.352 | 1.347 | 0.006 | 0.008 | 44 | 0.76 |
| C3D-C4D | 1.535 | 1.526 | 0.010 | 0.014 | 577 | 0.72 |
| C6A-N1A | 1.355 | 1.350 | 0.005 | 0.010 | 239 | 0.51 |
| C6A-N6A | 1.343 | 1.336 | 0.007 | 0.014 | 531 | 0.50 |
| O4B-C1B | 1.410 | 1.415 | -0.006 | 0.011 | 592 | 0.49 |
| O2B-C2B | 1.418 | 1.423 | -0.006 | 0.013 | 3264 | 0.43 |
| O4B-C4B | 1.448 | 1.444 | 0.005 | 0.011 | 1633 | 0.42 |
| C2A-N1A | 1.339 | 1.335 | 0.005 | 0.011 | 829 | 0.41 |
| C4A-N3A | 1.344 | 1.339 | 0.005 | 0.012 | 542 | 0.40 |
| C8A-N7A | 1.314 | 1.311 | 0.003 | 0.008 | 447 | 0.38 |
| PN-O3 | 1.598 | 1.594 | 0.004 | 0.011 | 21 | 0.38 |
| C2D-C3D | 1.526 | 1.530 | -0.005 | 0.013 | 510 | 0.36 |
| C3N-C7N | 1.507 | 1.501 | 0.006 | 0.016 | 409 | 0.36 |
| C2B-C3B | 1.526 | 1.530 | -0.005 | 0.013 | 510 | 0.36 |
| C8A-N9A | 1.372 | 1.370 | 0.003 | 0.008 | 190 | 0.35 |
| C5N-C4N | 1.388 | 1.384 | 0.004 | 0.010 | 2659 | 0.34 |
| C2N-N1N | 1.346 | 1.344 | 0.002 | 0.006 | 10 | 0.33 |
| PA-O3 | 1.591 | 1.594 | -0.003 | 0.011 | 21 | 0.31 |
| C2A-N3A | 1.338 | 1.335 | 0.003 | 0.011 | 829 | 0.30 |
| O4D-C1D | 1.407 | 1.404 | 0.004 | 0.012 | 17 | 0.29 |
| C2N-C3N | 1.384 | 1.381 | 0.002 | 0.007 | 79 | 0.28 |
| C5B-C4B | 1.504 | 1.508 | -0.003 | 0.012 | 1182 | 0.28 |
| C5A-C6A | 1.411 | 1.408 | 0.003 | 0.010 | 166 | 0.27 |
| O7N-C7N | 1.233 | 1.239 | -0.006 | 0.025 | 568 | 0.24 |
| O5D-C5D | 1.446 | 1.443 | 0.003 | 0.015 | 124 | 0.22 |
| PA-O1A | 1.478 | 1.482 | -0.004 | 0.018 | 31 | 0.20 |
| C5D-C4D | 1.510 | 1.508 | 0.002 | 0.012 | 1182 | 0.20 |
| C5A-N7A | 1.388 | 1.387 | 0.001 | 0.007 | 359 | 0.16 |
| PN-O1N | 1.479 | 1.482 | -0.002 | 0.018 | 31 | 0.12 |
| O2D-C2D | 1.425 | 1.423 | 0.001 | 0.013 | 3264 | 0.10 |
| C1B-N9A | 1.460 | 1.459 | 0.001 | 0.012 | 78 | 0.09 |
| O5B-C5B | 1.444 | 1.443 | 0.001 | 0.015 | 124 | 0.07 |
| C2D-C1D | 1.529 | 1.530 | -0.001 | 0.014 | 354 | 0.06 |
| C4A-N9A | 1.374 | 1.374 | -0.000 | 0.008 | 181 | 0.05 |
| C5A-C4A | 1.387 | 1.388 | -0.001 | 0.011 | 332 | 0.05 |
| C6N-C5N | 1.376 | 1.376 | -0.000 | 0.018 | 211 | 0.03 |
| O3B-C3B | 1.423 | 1.423 | 0.000 | 0.013 | 3264 | 0.01 |
Mogul angles schematic | ||||||
| atoms | actual in ° | Mogul mean in ° | difference in ° | Mogul σ in ° | Mogul # samples | Zscore |
|---|---|---|---|---|---|---|
| C4B-O4B-C1B | 105.7 | 109.5 | -3.8 | 1.4 | 434 | 2.61 |
| C2D-C1D-N1N | 117.8 | 113.6 | 4.2 | 1.8 | 20 | 2.30 |
| O4B-C1B-N9A | 111.3 | 108.4 | 2.9 | 1.3 | 66 | 2.25 |
| O5B-C5B-C4B | 104.6 | 109.1 | -4.5 | 2.3 | 84 | 1.93 |
| C3D-C2D-C1D | 99.4 | 101.5 | -2.1 | 1.2 | 285 | 1.82 |
| O4B-C1B-C2B | 104.4 | 106.5 | -2.1 | 1.2 | 307 | 1.82 |
| C4N-C3N-C7N | 117.2 | 120.7 | -3.5 | 2.4 | 598 | 1.45 |
| C3B-C2B-C1B | 99.8 | 101.5 | -1.7 | 1.2 | 273 | 1.45 |
| O3D-C3D-C2D | 108.6 | 111.9 | -3.3 | 2.6 | 938 | 1.26 |
| C4A-N9A-C1B | 125.0 | 127.0 | -1.9 | 1.8 | 67 | 1.10 |
| O4D-C1D-C2D | 105.9 | 107.0 | -1.1 | 1.1 | 17 | 1.07 |
| O3-PA-O1A | 111.2 | 108.7 | 2.5 | 2.4 | 19 | 1.02 |
| O4B-C4B-C3B | 106.6 | 105.3 | 1.3 | 1.3 | 448 | 1.02 |
| O7N-C7N-C3N | 118.7 | 119.5 | -0.8 | 0.8 | 373 | 1.00 |
| C1B-N9A-C8A | 128.6 | 126.8 | 1.8 | 1.9 | 65 | 0.96 |
| O2D-C2D-C1D | 113.2 | 110.6 | 2.6 | 2.8 | 336 | 0.93 |
| C2B-C1B-N9A | 115.5 | 114.1 | 1.4 | 1.5 | 59 | 0.92 |
| C6N-C5N-C4N | 119.2 | 119.9 | -0.7 | 0.7 | 149 | 0.92 |
| C6N-N1N-C2N | 121.8 | 122.2 | -0.3 | 0.4 | 10 | 0.88 |
| C3N-C7N-N7N | 118.7 | 117.9 | 0.8 | 1.0 | 373 | 0.84 |
| C4D-O4D-C1D | 108.4 | 109.5 | -1.1 | 1.4 | 447 | 0.80 |
| O4D-C4D-C3D | 106.1 | 105.3 | 0.9 | 1.3 | 448 | 0.67 |
| O3D-C3D-C4D | 109.3 | 111.0 | -1.7 | 2.6 | 541 | 0.66 |
| C5N-C4N-C3N | 119.9 | 120.4 | -0.4 | 0.7 | 3664 | 0.66 |
| C2B-C3B-C4B | 103.1 | 102.5 | 0.6 | 1.0 | 376 | 0.64 |
| O3B-C3B-C2B | 110.3 | 111.9 | -1.6 | 2.6 | 938 | 0.63 |
| C2N-C3N-C4N | 118.4 | 118.8 | -0.4 | 0.7 | 35 | 0.55 |
| O3B-C3B-C4B | 112.3 | 111.0 | 1.3 | 2.6 | 541 | 0.51 |
| C6N-N1N-C1D | 118.3 | 119.3 | -1.0 | 2.1 | 10 | 0.48 |
| O7N-C7N-N7N | 122.2 | 122.8 | -0.6 | 1.3 | 516 | 0.45 |
| C3N-C2N-N1N | 120.4 | 120.1 | 0.2 | 0.5 | 24 | 0.44 |
| C5B-C4B-C3B | 114.5 | 115.3 | -0.8 | 1.8 | 70 | 0.44 |
| O5D-C5D-C4D | 108.1 | 109.1 | -0.9 | 2.3 | 84 | 0.39 |
| C2A-N1A-C6A | 118.9 | 118.5 | 0.4 | 1.0 | 154 | 0.38 |
| O4D-C4D-C5D | 110.1 | 109.6 | 0.6 | 1.5 | 204 | 0.37 |
| O4B-C4B-C5B | 110.1 | 109.6 | 0.5 | 1.5 | 204 | 0.36 |
| O4D-C1D-N1N | 108.6 | 108.4 | 0.2 | 0.7 | 16 | 0.34 |
| O2B-C2B-C3B | 112.7 | 111.9 | 0.8 | 2.6 | 938 | 0.29 |
| O3-PN-O1N | 109.4 | 108.7 | 0.7 | 2.4 | 19 | 0.29 |
| N3A-C4A-N9A | 127.4 | 127.1 | 0.3 | 1.2 | 126 | 0.28 |
| C6A-C5A-N7A | 132.4 | 132.1 | 0.3 | 1.2 | 118 | 0.27 |
| O2B-C2B-C1B | 111.3 | 110.6 | 0.7 | 2.8 | 323 | 0.24 |
| N6A-C6A-N1A | 118.5 | 118.2 | 0.3 | 1.2 | 214 | 0.24 |
| C5A-C4A-N9A | 105.9 | 105.7 | 0.1 | 0.5 | 148 | 0.23 |
| C5D-C4D-C3D | 114.9 | 115.3 | -0.4 | 1.8 | 70 | 0.23 |
| C2A-N3A-C4A | 111.0 | 111.5 | -0.4 | 2.0 | 224 | 0.22 |
| C2N-N1N-C1D | 119.8 | 119.3 | 0.5 | 2.1 | 10 | 0.22 |
| O2D-C2D-C3D | 111.4 | 111.9 | -0.6 | 2.6 | 938 | 0.22 |
| C6A-C5A-C4A | 116.9 | 117.0 | -0.1 | 0.7 | 127 | 0.20 |
| C5A-C6A-N6A | 123.8 | 123.7 | 0.2 | 1.0 | 144 | 0.15 |
| C5A-C4A-N3A | 126.8 | 126.8 | -0.1 | 0.7 | 212 | 0.13 |
| C2D-C3D-C4D | 102.4 | 102.5 | -0.1 | 1.0 | 376 | 0.11 |
| C5N-C6N-N1N | 120.1 | 120.2 | -0.1 | 0.9 | 38 | 0.10 |
| N3A-C2A-N1A | 128.7 | 128.8 | -0.1 | 0.9 | 329 | 0.09 |
| C4A-C5A-N7A | 110.7 | 110.7 | 0.0 | 0.5 | 241 | 0.06 |
| C4A-N9A-C8A | 105.7 | 105.7 | -0.0 | 0.5 | 150 | 0.05 |
| C5A-C6A-N1A | 117.6 | 117.6 | 0.0 | 0.9 | 133 | 0.04 |
| C5A-N7A-C8A | 103.7 | 103.7 | -0.0 | 0.5 | 265 | 0.03 |
| N9A-C8A-N7A | 114.1 | 114.1 | -0.0 | 0.7 | 173 | 0.02 |
| PN-O3-PA | 132.5 | 132.5 | 0.0 | 3.0 | 21 | 0.01 |
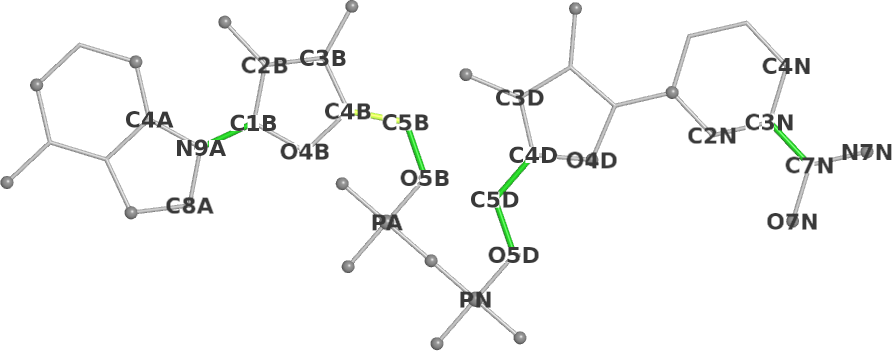 Mogul dihedrals schematic | ||||
| atoms | actual torsion angle in ° | Mogul histogram | Mogul # samples | classification or % Mogul population within ± 10° |
|---|---|---|---|---|
| …-C1B-N9A-… | common | |||
| O4B-C1B-N9A-C8A | 71.1 | 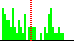 | 64 | 12% |
| O4B-C1B-N9A-C4A | -119.9 | 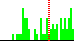 | 68 | 16% |
| C2B-C1B-N9A-C4A | 121.4 | 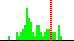 | 66 | 18% |
| C2B-C1B-N9A-C8A | -47.7 | 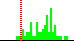 | 63 | 11% |
| …-C3N-C7N-… | common | |||
| C4N-C3N-C7N-O7N | -19.1 |  | 707 | 39% |
| C2N-C3N-C7N-N7N | -12.0 |  | 111 | 42% |
| C2N-C3N-C7N-O7N | 160.6 |  | 111 | 43% |
| C4N-C3N-C7N-N7N | 168.4 |  | 707 | 40% |
| …-C4B-C5B-… | rare | |||
| C3B-C4B-C5B-O5B | -67.1 | 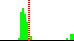 | 63 | 40% |
| O4B-C4B-C5B-O5B | 172.8 | 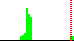 | 63 | 6% |
| …-C4D-C5D-… | common | |||
| C3D-C4D-C5D-O5D | 54.8 | 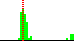 | 63 | 84% |
| O4D-C4D-C5D-O5D | -65.0 | 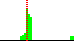 | 63 | 90% |
| …-C5B-O5B-… | common | |||
| C4B-C5B-O5B-PA | 128.4 | 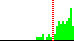 | 74 | 14% |
| …-C5D-O5D-… | common | |||
| C4D-C5D-O5D-PN | 147.5 | 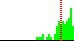 | 74 | 42% |
Mogul rings schematic | ||
| atoms | Mogul # samples | Ring strangeness score‡ in ° |
|---|---|---|
| C4B-O4B-C1B-C2B-C3B | 191 | 1.4 |
| C4D-O4D-C1D-C2D-C3D | 191 | 0.5 |
| C5A-C6A-N1A-C2A-N3A-C4A | 184 | 0.2 |
| N9A-C8A-N7A-C5A-C4A | 186 | 0.1 |
‡ 'ring strangeness score' is the RMS difference in torsion angles between the instance of the ring in the ligand in the model, and the nearest instance that mogul finds in the CSD.