Electron density around 880 A 501 :
2mFo-DFc (at 0.7 rmsd) in grey.
mFo-DFc (at 3 rmsd) in red (negative) and green (positive),
(picture produced with PyMOL)
Electron density around 880 A 501 : |
| Database ID | 880 (PDB) |
| 3-letter code | 880 |
| CC(2mFo-DFc) | 0.9557 |
| min(B-factor)‡ | 28.7 |
| avg(B-factor)‡ | 36.1 |
| max(B-factor)‡ | 44.2 |
| min(occupancy)‡ | 1.00 |
| max(occupancy)‡ | 1.00 |
| ‡ hydrogen atoms excluded | |
| Mogul Analysis: | |
|---|---|
| 'bad' bonds | 0/38 |
| 'bad' bond angles | 0/43 |
| 'unusual' dihedrals | 0/4 |
| 'bad' rings | 0/4 |
| bonds rms Z | 0.554 |
| angles rms Z | 1.028 |
Mogul bonds schematic | ||||||
| atoms | actual in Å | Mogul mean in Å | difference in Å | Mogul σ in Å | Mogul # samples | Zscore |
|---|---|---|---|---|---|---|
| C38-CL45 | 1.718 | 1.734 | -0.016 | 0.012 | 3693 | 1.34 |
| C5-C4 | 1.392 | 1.377 | 0.015 | 0.013 | 125 | 1.09 |
| C39-C38 | 1.375 | 1.388 | -0.013 | 0.013 | 403 | 0.98 |
| C2-N3 | 1.302 | 1.316 | -0.014 | 0.015 | 17 | 0.95 |
| C50-N51 | 1.354 | 1.341 | 0.012 | 0.013 | 434 | 0.95 |
| C36-C5 | 1.484 | 1.478 | 0.006 | 0.007 | 10 | 0.88 |
| C7-C6 | 1.532 | 1.514 | 0.018 | 0.023 | 243 | 0.78 |
| C58-C57 | 1.528 | 1.516 | 0.012 | 0.018 | 1743 | 0.71 |
| C50-N54 | 1.350 | 1.342 | 0.008 | 0.013 | 12 | 0.67 |
| C59-C58 | 1.527 | 1.516 | 0.011 | 0.018 | 1743 | 0.62 |
| C5-N1 | 1.398 | 1.389 | 0.008 | 0.014 | 79 | 0.59 |
| C55-N54 | 1.466 | 1.458 | 0.008 | 0.013 | 47 | 0.59 |
| C4-N3 | 1.377 | 1.385 | -0.008 | 0.013 | 288 | 0.58 |
| C56-C55 | 1.509 | 1.518 | -0.008 | 0.015 | 793 | 0.55 |
| C21-C20 | 1.520 | 1.512 | 0.008 | 0.014 | 29 | 0.55 |
| C6-N1 | 1.472 | 1.467 | 0.005 | 0.012 | 575 | 0.42 |
| C10-C7 | 1.492 | 1.509 | -0.017 | 0.040 | 1023 | 0.42 |
| C20-N19 | 1.474 | 1.467 | 0.007 | 0.018 | 139 | 0.42 |
| C50-N49 | 1.345 | 1.340 | 0.004 | 0.011 | 146 | 0.39 |
| C41-C35 | 1.396 | 1.391 | 0.005 | 0.013 | 4266 | 0.39 |
| C17-C18 | 1.517 | 1.512 | 0.005 | 0.014 | 29 | 0.36 |
| C40-C39 | 1.388 | 1.384 | 0.004 | 0.013 | 2673 | 0.31 |
| C41-C40 | 1.385 | 1.382 | 0.003 | 0.010 | 4486 | 0.30 |
| C18-N19 | 1.472 | 1.467 | 0.005 | 0.018 | 139 | 0.29 |
| C60-C55 | 1.514 | 1.518 | -0.004 | 0.015 | 793 | 0.26 |
| C36-N51 | 1.343 | 1.341 | 0.002 | 0.007 | 892 | 0.24 |
| C39-CL46 | 1.737 | 1.734 | 0.003 | 0.012 | 3693 | 0.23 |
| C37-C38 | 1.383 | 1.381 | 0.002 | 0.009 | 2188 | 0.20 |
| C48-N49 | 1.341 | 1.340 | 0.002 | 0.011 | 2774 | 0.17 |
| C35-C4 | 1.479 | 1.478 | 0.002 | 0.010 | 913 | 0.16 |
| C21-C16 | 1.532 | 1.531 | 0.001 | 0.014 | 1072 | 0.09 |
| C17-C16 | 1.530 | 1.531 | -0.001 | 0.014 | 1072 | 0.08 |
| C47-C36 | 1.390 | 1.389 | 0.001 | 0.010 | 1673 | 0.08 |
| C57-C56 | 1.526 | 1.525 | 0.001 | 0.013 | 1827 | 0.07 |
| C59-C60 | 1.524 | 1.525 | -0.001 | 0.013 | 1827 | 0.07 |
| C47-C48 | 1.380 | 1.381 | -0.001 | 0.011 | 2637 | 0.07 |
| C37-C35 | 1.391 | 1.391 | -0.000 | 0.009 | 1816 | 0.05 |
| C2-N1 | 1.375 | 1.375 | 0.000 | 0.013 | 11 | 0.00 |
Mogul angles schematic | ||||||
| atoms | actual in ° | Mogul mean in ° | difference in ° | Mogul σ in ° | Mogul # samples | Zscore |
|---|---|---|---|---|---|---|
| C5-C36-N51 | 118.5 | 115.8 | 2.7 | 1.0 | 10 | 2.62 |
| C56-C55-N54 | 114.8 | 110.6 | 4.3 | 1.7 | 34 | 2.49 |
| C60-C55-N54 | 114.4 | 110.6 | 3.8 | 1.7 | 34 | 2.24 |
| N54-C50-N51 | 119.2 | 116.5 | 2.7 | 1.4 | 22 | 1.95 |
| C35-C4-C5 | 132.1 | 130.0 | 2.0 | 1.3 | 92 | 1.54 |
| C47-C36-C5 | 119.5 | 121.1 | -1.6 | 1.1 | 10 | 1.45 |
| C40-C39-CL46 | 120.2 | 118.4 | 1.9 | 1.3 | 2376 | 1.42 |
| C35-C4-N3 | 117.3 | 119.3 | -2.0 | 1.4 | 227 | 1.39 |
| N54-C50-N49 | 114.6 | 116.5 | -1.9 | 1.4 | 22 | 1.38 |
| C47-C36-N51 | 122.0 | 123.0 | -1.0 | 0.8 | 691 | 1.22 |
| C39-C38-CL45 | 119.9 | 120.9 | -1.0 | 0.8 | 728 | 1.16 |
| C38-C39-CL46 | 120.0 | 120.9 | -0.9 | 0.8 | 728 | 1.02 |
| C4-C5-N1 | 104.5 | 105.4 | -0.9 | 0.9 | 25 | 1.02 |
| C6-N1-C2 | 125.6 | 127.1 | -1.6 | 1.6 | 26 | 1.00 |
| C59-C58-C57 | 111.8 | 110.9 | 0.9 | 1.0 | 1269 | 0.87 |
| C6-N1-C5 | 127.8 | 126.6 | 1.2 | 1.4 | 14 | 0.86 |
| C48-C47-C36 | 116.5 | 117.1 | -0.6 | 0.7 | 16 | 0.86 |
| C47-C48-N49 | 123.4 | 124.1 | -0.7 | 0.9 | 427 | 0.81 |
| C37-C38-C39 | 120.9 | 120.2 | 0.7 | 1.0 | 476 | 0.73 |
| C20-N19-C18 | 111.4 | 110.5 | 0.9 | 1.3 | 60 | 0.67 |
| C18-C17-C16 | 109.0 | 109.8 | -0.9 | 1.5 | 90 | 0.59 |
| N49-C50-N51 | 126.0 | 126.6 | -0.5 | 1.0 | 20 | 0.57 |
| C59-C60-C55 | 110.7 | 111.2 | -0.5 | 0.8 | 586 | 0.57 |
| C10-C7-C6 | 113.3 | 111.8 | 1.4 | 2.5 | 217 | 0.57 |
| C58-C57-C56 | 111.8 | 111.3 | 0.5 | 1.0 | 1421 | 0.56 |
| C41-C40-C39 | 119.8 | 120.2 | -0.4 | 0.7 | 492 | 0.53 |
| C37-C38-CL45 | 119.1 | 118.4 | 0.7 | 1.2 | 1948 | 0.53 |
| C4-N3-C2 | 106.0 | 105.2 | 0.7 | 1.6 | 12 | 0.47 |
| C36-N51-C50 | 116.3 | 116.7 | -0.4 | 0.9 | 14 | 0.44 |
| C37-C35-C4 | 120.5 | 119.9 | 0.6 | 1.4 | 31 | 0.43 |
| C41-C35-C37 | 119.1 | 119.4 | -0.3 | 0.9 | 2010 | 0.34 |
| C40-C41-C35 | 120.6 | 120.8 | -0.2 | 0.7 | 3513 | 0.33 |
| C48-N49-C50 | 115.3 | 115.6 | -0.2 | 0.7 | 106 | 0.32 |
| C35-C37-C38 | 119.9 | 120.1 | -0.2 | 0.7 | 61 | 0.23 |
| C20-C21-C16 | 109.5 | 109.8 | -0.3 | 1.5 | 90 | 0.21 |
| C57-C56-C55 | 111.0 | 111.2 | -0.2 | 0.8 | 586 | 0.21 |
| C5-C4-N3 | 110.5 | 110.3 | 0.2 | 1.2 | 77 | 0.20 |
| C21-C16-C17 | 109.3 | 109.5 | -0.2 | 1.2 | 259 | 0.19 |
| C41-C35-C4 | 120.4 | 120.7 | -0.3 | 1.7 | 1605 | 0.19 |
| C60-C55-C56 | 110.8 | 110.7 | 0.1 | 0.8 | 315 | 0.17 |
| C7-C6-N1 | 112.6 | 112.5 | 0.2 | 1.9 | 33 | 0.10 |
| C40-C39-C38 | 119.7 | 119.7 | 0.0 | 0.8 | 229 | 0.03 |
| C58-C59-C60 | 111.3 | 111.3 | 0.0 | 1.0 | 1421 | 0.03 |
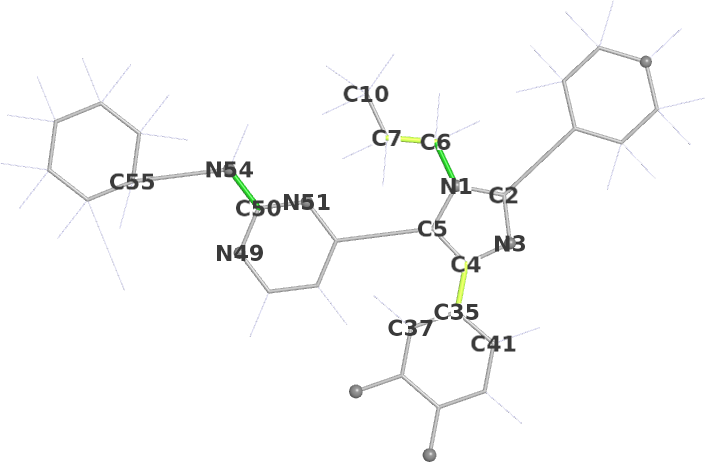 Mogul dihedrals schematic | ||||
| atoms | actual torsion angle in ° | Mogul histogram | Mogul # samples | classification or % Mogul population within ± 10° |
|---|---|---|---|---|
| …-C35-C4-… | rare | |||
| C41-C35-C4-C5 | -129.6 | 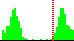 | 452 | 6% |
| C37-C35-C4-N3 | -130.9 | 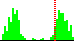 | 528 | 19% |
| C41-C35-C4-N3 | 46.6 | 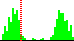 | 528 | 16% |
| C37-C35-C4-C5 | 52.9 | 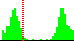 | 452 | 9% |
| …-C50-N54-… | common | |||
| N51-C50-N54-C55 | -7.8 |  | 22 | 45% |
| N49-C50-N54-C55 | 167.4 |  | 22 | 55% |
| …-C6-C7-… | rare | |||
| N1-C6-C7-C10 | 32.4 |  | 196 | 1% |
| …-C6-N1-… | common | |||
| C7-C6-N1-C5 | -108.2 | 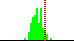 | 97 | 32% |
| C7-C6-N1-C2 | 71.6 | 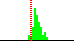 | 49 | 43% |
Mogul rings schematic | ||
| atoms | Mogul # samples | Ring strangeness score‡ in ° |
|---|---|---|
| C55-C56-C57-C58-C59-C60 | 148 | 0.5 |
| N1-C2-N3-C4-C5 | 58 | 0.1 |
| C36-C47-C48-N49-C50-N51 | 41 | 0.7 |
| C35-C37-C38-C39-C40-C41 | 246 | 0.1 |
‡ 'ring strangeness score' is the RMS difference in torsion angles between the instance of the ring in the ligand in the model, and the nearest instance that mogul finds in the CSD.